In a groundbreaking study, researchers have traced the evolutionary transition from sodium-coupled to proton-coupled glutamate transporters, shedding light on a pivotal shift in microbial protein function. This analysis, conducted through a sophisticated workflow of sequence retrieval and phylogenetic analysis, offers new insights into the biochemical evolution of these essential transporters.
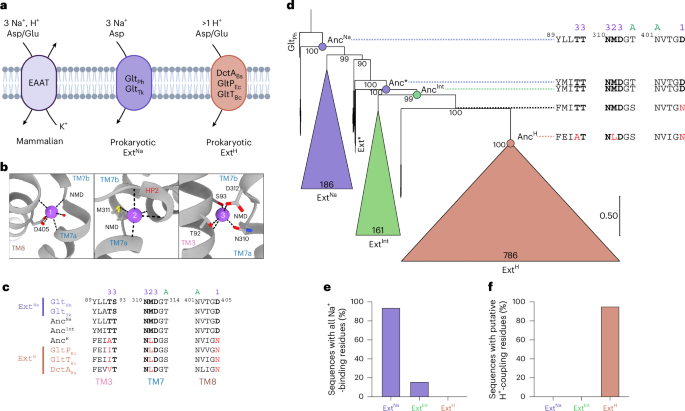
The study, which utilized maximum-likelihood (ML) phylogenies, aimed to capture the transition from Na+-coupled to H+-coupled transporters. This transition is marked by distinctive sequence substitutions in the Na+-binding sites, which are prevalent across various bacterial phyla. The findings suggest that the emergence of proton-coupled transporters (ExtH) was likely a result of a single-gene duplication event, followed by horizontal gene transfer.
Tracing the Evolutionary Path
The researchers embarked on this study by retrieving a substantial dataset of 20,000 nonredundant sequences from the Bacillota/Firmicutes phylum, where ExtH sequences are predominantly found. Using advanced bioinformatics tools like PSI-BLAST and CD-HIT, these sequences were meticulously curated and aligned. The final sequence set comprised 1,410 sequences, which were then analyzed to construct a maximum-parsimony tree.
According to the study, the ancestral protein reconstruction (APR) revealed that the divergence into sodium-independent exchangers and subsequent proton-coupled transporters occurred after the split between bacteria and archaea. This finding underscores the complexity and adaptability of microbial life forms in response to environmental pressures.
Insights from Phylogenetic Analysis
Utilizing IQ-TREE software, the researchers generated ML trees and ancestral sequences, employing the LG + F + R protein evolution model. This model considers site-specific differences in evolutionary rates, providing a nuanced understanding of protein evolution. The study’s results indicate that the origin of glutamate transport was sodium-coupled, with subsequent adaptations leading to the development of proton-coupled transporters.
Notably, the Firmicutes tree yielded robust bootstrap values, indicating a high level of confidence in the phylogenetic analysis. In contrast, the Proteobacteria phylum presented challenges, with lower reproducibility in tree structures, highlighting the variability in evolutionary pathways across different bacterial groups.
Implications and Future Directions
The implications of this study extend beyond academic curiosity, offering potential applications in biotechnology and medicine. Understanding the evolutionary mechanisms behind transporter function could inform the development of novel antibiotics or the engineering of microorganisms for industrial processes.
As the researchers continue to explore the evolutionary dynamics of microbial proteins, this study serves as a foundation for future investigations into the genetic and biochemical pathways that have shaped life on Earth. The integration of advanced bioinformatics tools and experimental techniques will undoubtedly propel further discoveries in this rapidly evolving field.
In conclusion, the evolutionary analysis of sodium coupling in glutamate transporters not only enhances our understanding of microbial evolution but also opens new avenues for scientific exploration and innovation.







