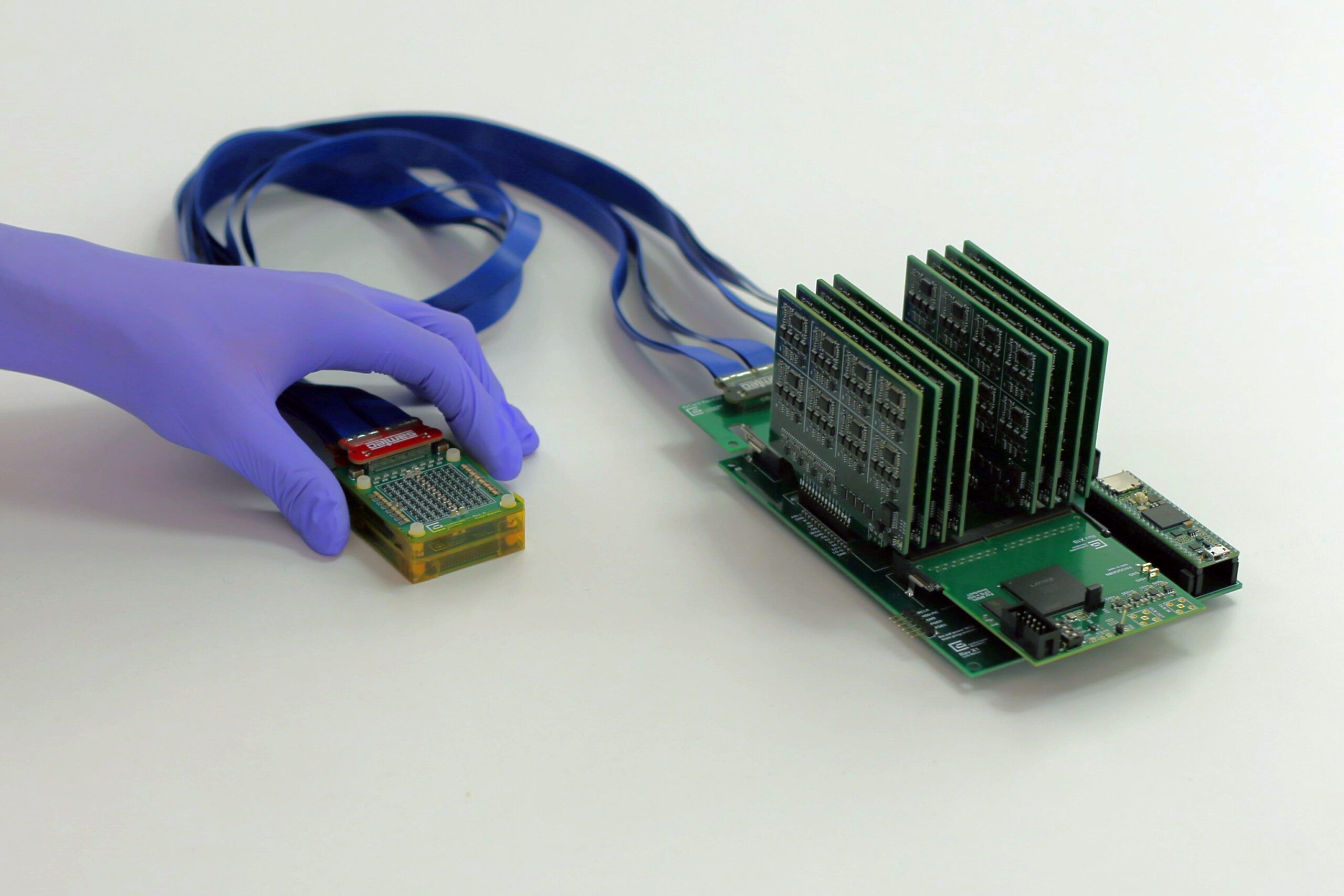
New York, NY [October 1, 2025]—A groundbreaking software platform, Giotto Suite, has been developed by researchers at the Icahn School of Medicine at Mount Sinai in New York, Boston Medical Center, and Boston University Chobanian & Avedisian School of Medicine. This innovative tool aims to streamline the analysis of molecular structures within tissues, providing a significant leap forward in both healthy and disease state research. The details were published in the October 1 online issue of Nature Methods [DOI: 10.1038/s41592-025-02817-w].
In recent years, the field of spatial omics has advanced dramatically, enabling scientists to map RNA and proteins within intact tissues with unprecedented detail. These technological strides are crucial for understanding cellular behavior and interactions within various tissue microenvironments, particularly in conditions such as cancer, neurodegenerative diseases, and immune disorders.
Addressing the Data Analysis Challenge
Despite the wealth of data generated by spatial omics technologies, the primary challenge remains in effectively analyzing and interpreting this complex information. “The biggest obstacle is no longer generating the information—it’s making sense of it,” said Dr. Guo-Cheng Yuan, co-senior corresponding author and Professor of Genetics and Genomic Sciences at the Icahn School of Medicine at Mount Sinai. “Biologists need tools that can keep pace with the growing complexity and scale of these datasets and help them draw meaningful conclusions.”
Giotto Suite was created to meet this need. It is a modular software suite built in R, a programming language widely used for data analysis. The tool is designed to be compatible with spatial data from any platform, supporting a broad range of analyses, including data integration across different types of measurements and resolutions. Its flexible structure allows users to begin analysis from multiple points, tailored to their specific research needs.
Future Developments and Community Engagement
While the current version of Giotto Suite includes only a subset of available analytical methods, the developers emphasize its potential for growth. The platform is intended to evolve, with plans to expand its features, enhance interoperability with other software packages, and provide ongoing training and support to the research community.
The research teams plan to apply Giotto Suite in their own studies of cancer and neurodegenerative diseases. They are also committed to refining the tool based on feedback from the scientific community. The software is freely accessible to researchers at giottosuite.com, with the hope that it will encourage broader adoption of spatial omics technologies in biomedical research.
Support and Collaboration
The study’s authors include Jiaji George Chen, Joselyn Cristina Chávez-Fuentes, Matthew O’Brien, Junxiang Xu, Edward Ruiz, Wen Wang, Iqra Amin, Jeffrey Sheridan, Sujung Crystal Shin, Sanjana Varada Hasyagar, Irzam Sarfraz, Pratishtha Guckhool, Adriana Sistig, Veronica Jarzabek, Guo-Cheng Yuan, and Ruben Dries. This work was supported by the Chan Zuckerberg Initiative’s Essential Open Source Software for Science Program (2022-252544), The Crazy 8 Initiative from the Alex’s Lemonade Stand Foundation for Childhood Cancer, and the National Institutes of Health (NIH) RF1MH128970, RF1MH133703, R01AG066028.
The announcement of Giotto Suite represents a significant step forward in the field of spatial omics, offering a versatile and powerful tool for researchers worldwide. As the platform continues to develop, it promises to enhance the capabilities of scientists in understanding and combating complex diseases.