The escalating problem of antibiotic-resistant bacteria has left health experts increasingly concerned. As bacteria evolve to withstand antibiotics, the world finds itself in a daunting arms race against these microscopic adversaries. In a groundbreaking study, researchers from the University of Tokyo have uncovered two distinct strategies employed by bacterial plasmids to disseminate antimicrobial resistance. This discovery, achieved through extensive data analysis, could pave the way for improved risk assessment and targeted laboratory experiments.
Bacteria, including those that cause diseases, are notorious for their rapid evolution. This agility is largely due to their ability to exchange genetic material, a process facilitated by plasmids—small DNA loops that bacteria can eject. Some plasmids can traverse a broad spectrum of bacterial species, while others remain confined to a select few. Given that plasmids often harbor genes for antimicrobial resistance, understanding their spread is vital for public health research.
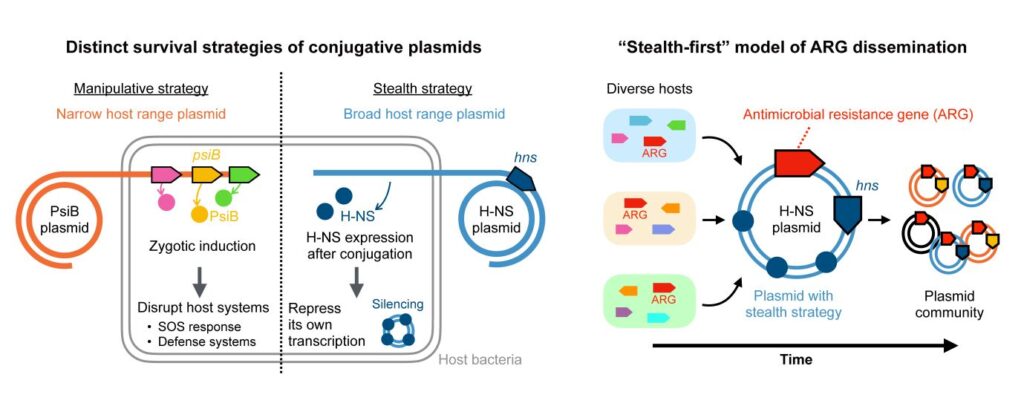
Unveiling Plasmid Survival Strategies
Ryuichi Ono, an undergraduate student from the Department of Bioinformatics and Systems Biology, explained the findings:
“Our latest work shows that plasmids follow two very different survival strategies: Some behave stealthily, keeping their genes mostly silent to minimize their impacts, while others act in a manipulative way, actively interfering with the host’s systems to secure their own survival.”
By analyzing over 10,000 plasmid sequences, the research team identified a clear pattern. Stealth plasmids tend to acquire new antibiotic-resistance genes first, which are then spread rapidly by manipulative plasmids—a process they term “stealth-first.”
Ono’s team focused on bacteria from the Enterobacterales group, which includes E. coli. They found that the stealth and manipulation strategies rarely coexist in the same plasmid. Moreover, plasmids employing either strategy typically carry more antibiotic-resistance genes than those using neither.
Genetic Signatures and Predictive Models
The research identified specific genes, hns for stealth plasmids and psiB for manipulative ones, which differentiate less selective plasmids from more selective ones. This led to the introduction of “plasmid survival strategies” as a framework for understanding plasmid evolution. Applying this framework to 48 major antibiotic-resistance genes revealed a consistent pattern, offering insights into past resistance outbreaks and potential future ones.
The implications of these findings are significant. The research suggests that monitoring genes primarily found on stealth plasmids could serve as an early warning system for emerging resistance threats.
“Our stealth-first model suggests we may be able to anticipate future resistance threats by monitoring which genes appear mainly on stealth plasmids,”
said Naoki Konno, a graduate student at the Department of Biological Sciences. This model could become a vital tool in predicting the movement of resistance genes through bacterial populations.
Future Directions and Challenges
While the research offers a promising new model for understanding plasmid evolution, it is not without limitations. The study focused on a single bacterial group and was based on computational analyses. Future research across additional bacterial lineages, coupled with experimental validation, will be crucial to expand these findings.
The potential to use the stealth-first framework as an early warning system excites researchers worldwide. Many are striving to predict how resistance genes spread through bacterial populations, and the University of Tokyo’s work represents a significant step toward that goal. However, the path forward is challenging, requiring continued collaboration and innovation.
As the battle against antibiotic resistance intensifies, the insights gained from this study could prove invaluable. By understanding the mechanisms behind plasmid behavior, scientists hope to develop more effective strategies to combat the spread of resistance, ultimately safeguarding global health.