The growing threat of antibiotic-resistant bacteria has become a pressing concern for health experts worldwide. As these bacteria evolve and adapt to our current methods of control, particularly antibiotics, the situation resembles an arms race that humanity appears to be losing. In a groundbreaking study, researchers, including those from the University of Tokyo, have identified two distinct strategies employed by bacterial plasmids to disseminate antimicrobial resistance. This discovery, facilitated by extensive data analysis, could significantly enhance our ability to predict and mitigate future resistance threats.
For the first time, scientists have utilized computational analysis to uncover how bacterial plasmids, which are small loops of DNA, contribute to the rapid adaptation of bacteria, including pathogenic strains. These plasmids are known to carry genes that confer resistance to antibiotics, making their study crucial. Some plasmids can transfer across various bacterial species, while others are more restricted. Understanding their spread is vital for developing effective countermeasures against antibiotic resistance.
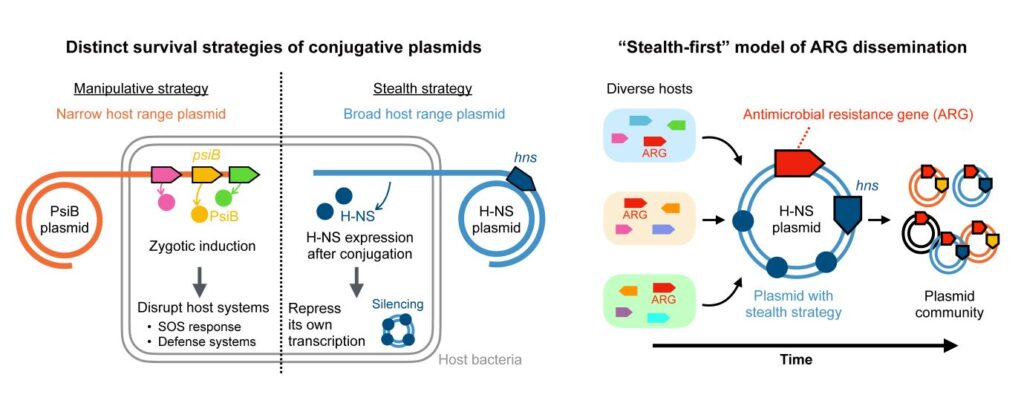
Unveiling the Dual Strategies of Plasmids
According to Ryuichi Ono, an undergraduate student from the University of Tokyo’s Department of Bioinformatics and Systems Biology, plasmids employ two distinct survival strategies: stealth and manipulation. Ono explains,
“Our latest work shows that plasmids follow two very different survival strategies: Some behave stealthily, keeping their genes mostly silent to minimize their impacts, while others act in a manipulative way, actively interfering with the host’s systems to secure their own survival.”
By analyzing over 10,000 plasmid sequences, Ono and his team discerned a clear pattern. Stealth plasmids tend to acquire new antibiotic-resistance genes first, while manipulative plasmids facilitate their rapid dissemination.
Ono’s team focused their analysis on Enterobacterales, a group of bacteria that includes E. coli. They found that stealth and manipulation strategies rarely coexist within the same plasmid. Plasmids utilizing either strategy generally harbor more antibiotic-resistance genes than those employing neither strategy. This finding introduces the concept of “plasmid survival strategies” as a framework for understanding plasmid evolution.
Implications for Future Research and Public Health
The research team’s findings offer a novel model for understanding plasmid evolution and their connection to potential targets. This model could significantly improve the tracking and prediction of new antibiotic-resistant infections. The identification of specific genes, such as hns for stealth plasmids and psiB for manipulative ones, provides a promising avenue for future research aimed at preventing the spread of antimicrobial resistance.
However, the study’s focus on a single bacterial group and reliance on computational analyses highlight the need for further investigation. Future research should encompass additional bacterial lineages and include experimental validation to build on these findings.
A New Framework for Predicting Resistance
Naoki Konno, a graduate student at the Department of Biological Sciences, emphasizes the potential of the “stealth-first” model as an early warning system for resistance threats. Konno notes,
“Our stealth-first model suggests we may be able to anticipate future resistance threats by monitoring which genes appear mainly on stealth plasmids. If a resistance gene is still confined to stealth plasmids and has not yet moved on to manipulative plasmids, it may be on the verge of entering a rapid second phase of spread.”
This framework could serve as a practical tool for predicting how resistance genes traverse bacterial populations, a goal pursued by many research groups globally.
As the battle against antibiotic resistance intensifies, the insights gained from this study represent a significant step forward. While challenges remain, the potential to develop an effective early warning system for resistance threats is a pursuit worth undertaking. By enhancing our understanding of plasmid strategies, researchers are paving the way for more effective interventions in the fight against antibiotic-resistant bacteria.







