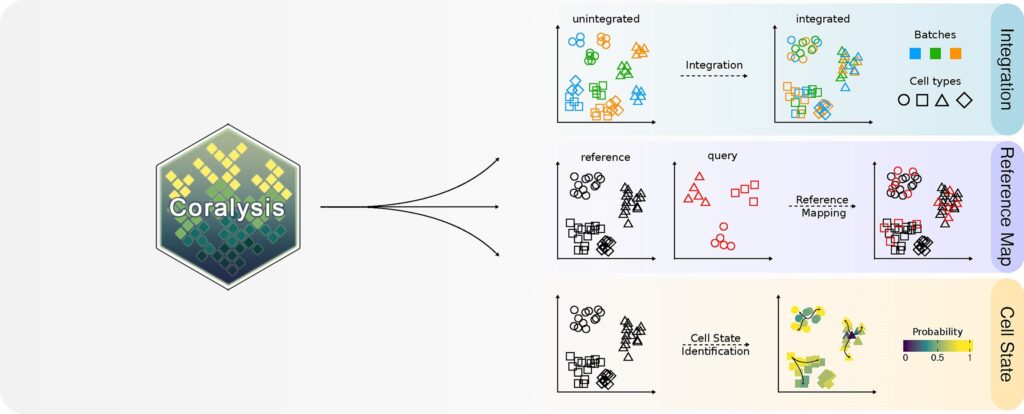
Researchers from the Turku Bioscience Center at the University of Turku, Finland, have unveiled a groundbreaking computational method designed to interpret complex single-cell data. This new tool aids scientists in identifying and categorizing cell types across various samples, marking a significant advancement in biomedical research.
The human body comprises approximately 37 trillion cells, each with unique characteristics despite their similarities. Modern single-cell technologies have revolutionized the ability to characterize this cellular heterogeneity by measuring numerous molecules, such as genes or proteins, across thousands of individual cells simultaneously. This capability provides profound insights into health and disease mechanisms.
Challenges in Data Integration
A single drop of blood contains billions of red blood cells and millions of immune cells, each with its own molecular “fingerprint.” Researchers utilize single-cell technologies in conjunction with computational methods to identify these unique patterns. However, integrating data from different samples poses a challenge, especially when cell types vary significantly or are present in disparate amounts. This often leads to errors in combining distinct cell types.
To address these challenges, scientists at the University of Turku have developed a machine learning-based algorithm capable of effectively integrating even imbalanced data across samples. This method, known as Coralysis, was developed at the Turku Bioscience Center under Professor Laura Elo’s Computational Biomedicine research group, which is also part of the InFLAMES Research Flagship.
“Single-cell technologies let us study the incredible diversity of cells, but comparing them across samples is tricky. This motivated us to develop a method to uncover these hidden patterns reliably,” says Associate Professor Sini Junttila.
Coralysis: A New Era in Data Analysis
Coralysis is likened to solving a complex puzzle, where pieces are grouped based on various features before being assembled. The algorithm employs multiple rounds of divisive clustering to progressively integrate cellular identities. António Sousa, the lead developer of Coralysis, explains that this method allows for the accurate prediction of cellular identities in new datasets and estimates the confidence of these predictions.
Implemented as open-source software, Coralysis is a machine learning tool that alleviates the cumbersome task of manually identifying cell types. Its unique ability to detect changing cellular states offers researchers a deeper understanding of complex single-cell data.
“Coralysis provides the scientific community with a new way to study cellular diversity and gain a deeper understanding of complex single-cell data. By making it openly available, we hope to support collaboration and accelerate discoveries across the global research community,” states Professor Laura Elo.
Implications for Future Research
The introduction of Coralysis could have far-reaching implications for biomedical research. By providing a more reliable method for data integration, researchers can explore cellular diversity with greater accuracy and confidence. This advancement is expected to accelerate discoveries and foster collaboration across the global scientific community.
As the field of single-cell analysis continues to evolve, tools like Coralysis will be instrumental in overcoming existing challenges and unlocking new possibilities in understanding cellular behaviors and disease mechanisms. The open-source nature of Coralysis ensures that researchers worldwide can access and contribute to its development, further enhancing its capabilities and applications.
Looking ahead, the team at the University of Turku plans to continue refining Coralysis and exploring its potential applications in various areas of biomedical research. This innovative tool represents a significant step forward in the quest to unravel the complexities of cellular data and its implications for human health.







